from frb_ml_utils import *
import frb_ml_utils
import numpy as np
import pandas as pd
import seaborn as sns
import scipy
import sklearn
import matplotlib.patches as patches
from matplotlib import pyplot as plt
from sklearn import svm
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier,AdaBoostClassifier
from sklearn.neighbors import NearestCentroid
from sklearn.metrics import confusion_matrix,ConfusionMatrixDisplay,accuracy_score,f1_score,roc_auc_score,balanced_accuracy_score,fbeta_score
from sklearn.model_selection import train_test_split
from sklearn.inspection import permutation_importance
from sklearn.preprocessing import StandardScaler
from imblearn.over_sampling import SMOTE
from imblearn.pipeline import Pipeline as imbpipeline
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from dtreeviz.trees import dtreeviz
from dtreeviz import clfvizCHIME = load_chime()
columns_to_use = ['bc_width','flux','fluence','dm_exc_ne2001',
'peak_freq',
'bright_temp','rest_width','freq_width','energy']
CHIME['bright_temp'] = np.log10(CHIME['bright_temp'])
CHIME['energy'] = np.log10(CHIME['energy'])
CHIME['rest_width'] = CHIME['rest_width'] * 1000
CHIME['bc_width'] = CHIME['bc_width'] * 1000
CHIME['freq_width'] = np.log10(CHIME['freq_width'])
chime_data = CHIME[columns_to_use]
chime_target = (CHIME['repeater_name'] != '-9999').to_numpy().astype('int')
X,test_X,y,test_y = train_test_split(chime_data,chime_target,test_size=0.3,stratify=chime_target)
scaler = StandardScaler()
scaler.fit(X)
X = scaler.transform(X)
test_X = scaler.transform(test_X)
chime_data = scaler.transform(chime_data)
X, y = SMOTE().fit_resample(X, y)2 78.8 0.00225301 FRB20180729A
12 101.5 0.00225301 FRB20180814A
38 101.0 0.00225301 FRB20180919A
49 94.7 0.00225301 FRB20180928A
75 101.3 0.00225301 FRB20181028A
76 101.3 0.00225301 FRB20181028A
77 101.3 0.00225301 FRB20181028A
78 101.3 0.00225301 FRB20181028A
79 101.3 0.00225301 FRB20181028A
81 62.3 0.00225301 FRB20181030A
82 62.5 0.00225301 FRB20181030B
158 83.6 0.00225301 FRB20181220A
174 92.6 0.00225301 FRB20181223C
221 96.1 0.00225301 FRB20190107B
399 100.8 0.00225301 FRB20190329A
459 79.4 0.00225301 FRB20190425A
571 100.7 0.00225301 FRB20190625E
572 100.7 0.00225301 FRB20190625E
573 100.7 0.00225301 FRB20190625E
576 101.5 0.00225301 FRB20190626ACHIME_for_plot = CHIME.copy()[columns_to_use]
CHIME_for_plot['Class'] = ['Repeating' if row['repeater_name'] != '-9999' else 'Non-repeating' for i,row in CHIME.iterrows()]
sns.set_theme('paper',"ticks",'dark')
sns.set_context("paper", rc={"font.size":10,"axes.labelsize":18})
p = sns.pairplot(pd.concat([CHIME_for_plot], axis=1), hue='Class')
for ax in p.axes.flat:
ax.tick_params(axis='both', labelsize=14)
p.legend.get_title().set_fontsize(15)
for legend_text in p.legend.get_texts():
legend_text.set_fontsize(15)
p.tight_layout()
# plt.savefig('./paper/features.pdf')<seaborn.axisgrid.PairGrid at 0x203f203c850>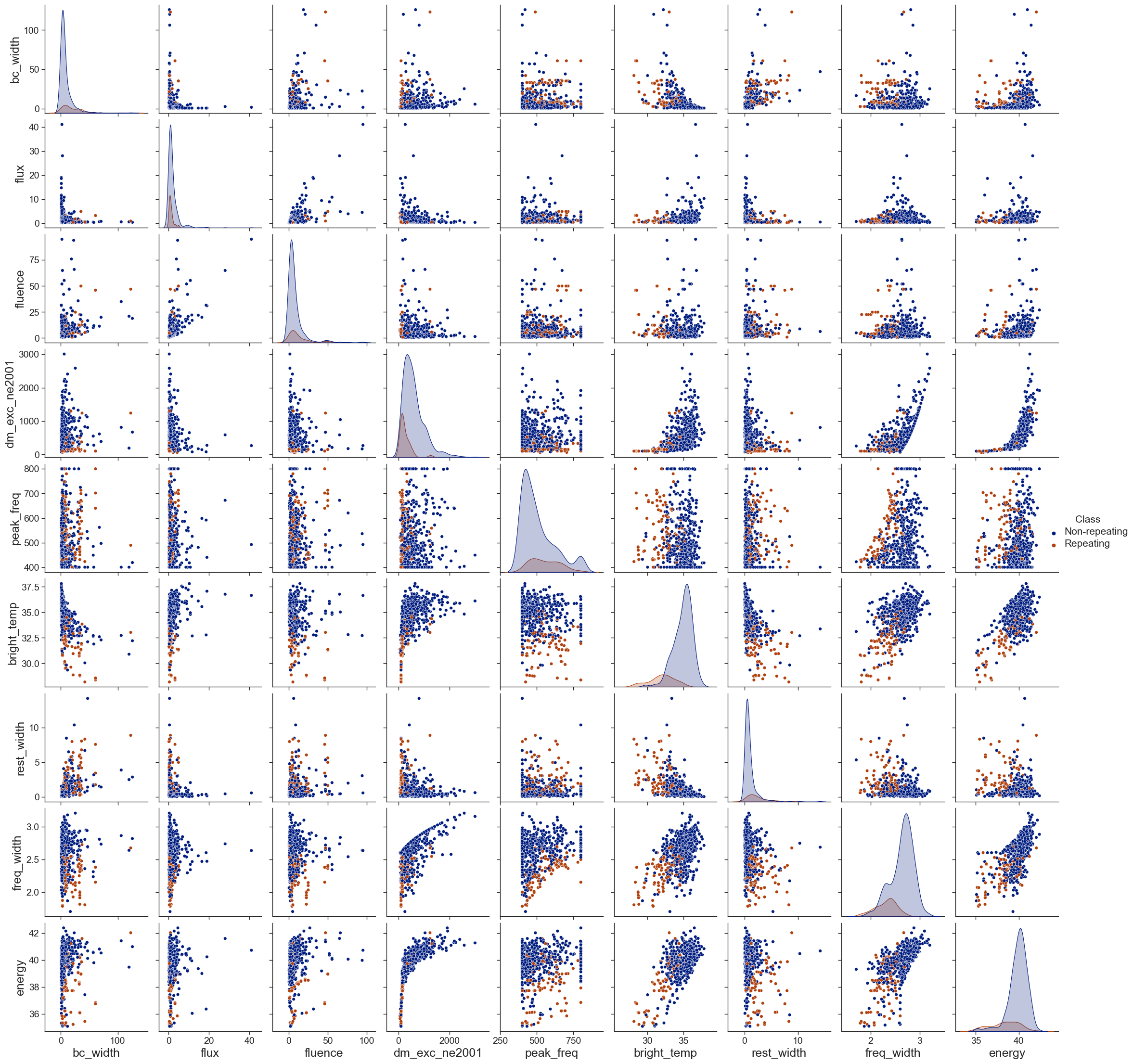
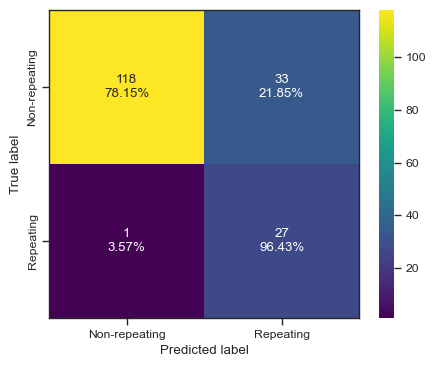
clf = svm.SVC()
clf.fit(X, y)
predictions = clf.predict(test_X)
print(predictions)
print(test_y)
print(accuracy_score(test_y,predictions))
print(f1_score(test_y,predictions))
print(roc_auc_score(test_y,predictions))
print(fbeta_score(test_y,predictions,beta=2))
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/svm_cm.pdf')[1 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 1 0 1 1 1 1 0 1 0 0 0
0 0 1 0 0 0 1 1 0 0 0 0 0 1 0 0 1 0 0 1 1 0 1 0 0 1 1 0 0 0 0 0 0 0 0 0 0
1 0 0 0 1 0 0 1 0 0 1 1 1 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1
0 0 0 1 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 1 0 1 1 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 0 0 0 0 0 0 0 1 0 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
0.8770949720670391
0.6944444444444445
0.8835146641438032
0.8012820512820514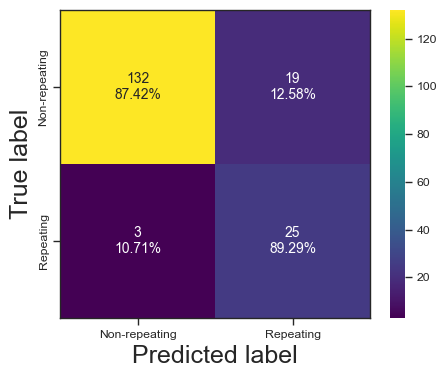
sns.set_theme('paper','ticks')
result = permutation_importance(clf, chime_data, chime_target, n_repeats=100, n_jobs=-1,scoring=f2_score)
forest_importances = pd.Series(result.importances_mean, index=columns_to_use)
fig, ax = plt.subplots()
forest_importances.plot.bar(yerr=result.importances_std, ax=ax,capsize=4)
ax.set_ylabel(r"Mean $F_2$ decrease")
fig.tight_layout()
plt.show()
# fig.savefig('./paper/svm_fi.pdf')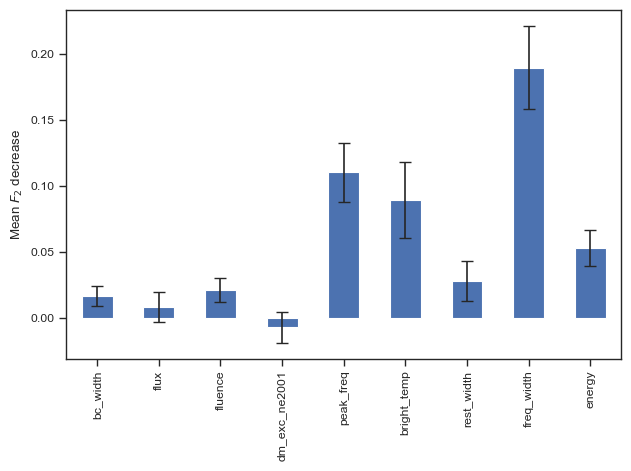
clf = RandomForestClassifier()
clf.fit(X, y)
predictions = clf.predict(test_X)
print(predictions)
print(test_y)
print(accuracy_score(test_y,predictions))
print(f1_score(test_y,predictions))
print(roc_auc_score(test_y,predictions))
print(fbeta_score(test_y,predictions,beta=2))
print(CHIME[np.logical_and(clf.predict(chime_data)==1, chime_target==0)]['tns_name'])
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/rf_cm.pdf')[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 1 0 1 1 1 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 1 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0
1 0 0 0 1 0 0 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 0 0 0 0 0 0 0 1 0 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
0.9385474860335196
0.8135593220338982
0.9053926206244087
0.8391608391608392
124 FRB20181128C
154 FRB20181218C
224 FRB20190109B
232 FRB20190112A
323 FRB20190218B
454 FRB20190423B
455 FRB20190423B
Name: tns_name, dtype: object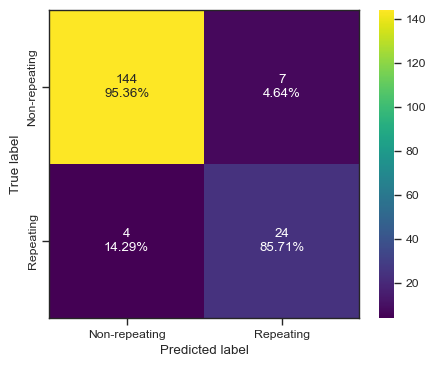
sns.set_theme('paper','ticks')
result = permutation_importance(clf, chime_data, chime_target, n_repeats=100, n_jobs=-1,scoring=f2_score)
forest_importances = pd.Series(result.importances_mean, index=columns_to_use)
fig, ax = plt.subplots()
forest_importances.plot.bar(yerr=result.importances_std, ax=ax,capsize=4)
ax.set_ylabel(r"Mean $F_2$ decrease")
fig.tight_layout()
plt.show()
# fig.savefig('./paper/rf_fi.pdf')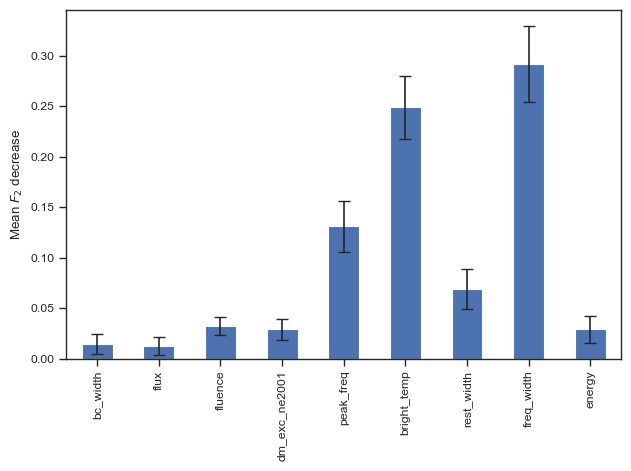
clf = AdaBoostClassifier()
clf.fit(X, y)
predictions = clf.predict(test_X)
print(predictions)
print(test_y)
print(accuracy_score(test_y,predictions))
print(f1_score(test_y,predictions))
print(roc_auc_score(test_y,predictions))
print(fbeta_score(test_y,predictions,beta=2))
print(CHIME[np.logical_and(clf.predict(chime_data)==1, chime_target==0)]['tns_name'])
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/ab_cm.pdf')[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 1 0 1 0 1 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 1 1 0 0 1 0 0 1 1 0 1 0 1 1 1 0 0 0 0 0 0 0 0 0 0
1 0 0 0 1 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 1
0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 1 1 1 1
0 0 0 1 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 1 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 0 0 0 0 0 0 0 1 0 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
0.8994413407821229
0.742857142857143
0.9113055818353832
0.8441558441558441
2 FRB20180729A
40 FRB20180920B
84 FRB20181030E
99 FRB20181118B
115 FRB20181125A
117 FRB20181125A
124 FRB20181128C
125 FRB20181128C
154 FRB20181218C
174 FRB20181223C
196 FRB20181229B
203 FRB20181231B
224 FRB20190109B
232 FRB20190112A
272 FRB20190129A
291 FRB20190206B
323 FRB20190218B
349 FRB20190228A
418 FRB20190409B
421 FRB20190410A
449 FRB20190422A
450 FRB20190422A
454 FRB20190423B
455 FRB20190423B
Name: tns_name, dtype: object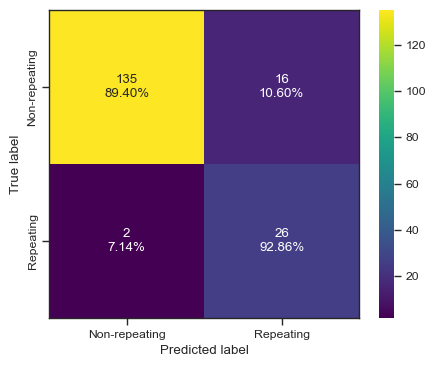
sns.set_theme('paper','ticks')
# result = permutation_importance(clf, test_X, test_y, n_repeats=100, n_jobs=-1,scoring=f2_score)
result = permutation_importance(clf, chime_data, chime_target, n_repeats=100, n_jobs=-1,scoring=f2_score)
forest_importances = pd.Series(result.importances_mean, index=columns_to_use)
fig, ax = plt.subplots()
forest_importances.plot.bar(yerr=result.importances_std, ax=ax,capsize=4)
ax.set_ylabel(r"Mean $F_2$ decrease")
fig.tight_layout()
plt.show()
# fig.savefig('./paper/ab_fi.pdf')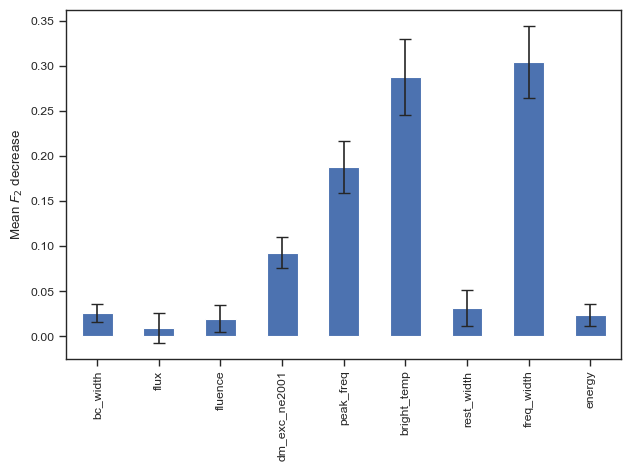
clf = NearestCentroid()
clf.fit(X, y)
predictions = clf.predict(test_X)
print(predictions)
print(test_y)
print(accuracy_score(test_y,predictions))
print(f1_score(test_y,predictions))
print(roc_auc_score(test_y,predictions))
print(fbeta_score(test_y,predictions,beta=2))
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/nc_cm.pdf')[0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 1 0 0 1 1 0 0 1 0 0 0
0 0 1 0 1 0 1 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 1 0 0 1 1 0 0 0 0 0 1 0 0 0 0
0 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1
0 0 0 0 0 0 0 0 0 0 1 1 0 0 1 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 1 1
0 0 0 1 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 1 0 0 1 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 0 0 0 0 0 0 0 1 0 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
0.8603351955307262
0.647887323943662
0.8444891201513718
0.7419354838709676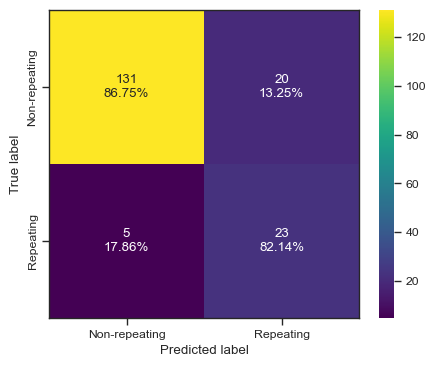
sns.set_theme('paper','ticks')
# result = permutation_importance(clf, test_X, test_y, n_repeats=100, n_jobs=-1,scoring=f2_score)
result = permutation_importance(clf, chime_data, chime_target, n_repeats=100, n_jobs=-1,scoring=f2_score)
forest_importances = pd.Series(result.importances_mean, index=columns_to_use)
fig, ax = plt.subplots()
forest_importances.plot.bar(yerr=result.importances_std, ax=ax,capsize=4)
ax.set_ylabel(r"Mean $F_2$ decrease")
fig.tight_layout()
plt.show()
# fig.savefig('./paper/nc_fi.pdf')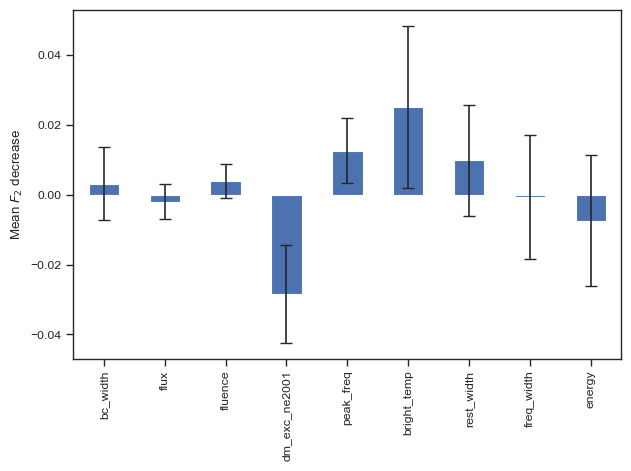
clf = LGBMClassifier()
clf.fit(X, y)
predictions = clf.predict(test_X)
print(predictions)
print(test_y)
print(accuracy_score(test_y,predictions))
print(f1_score(test_y,predictions))
print(roc_auc_score(test_y,predictions))
print(fbeta_score(test_y,predictions,beta=2))
print(CHIME[np.logical_and(clf.predict(chime_data)==1, chime_target==0)]['tns_name'])
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/lgbm_cm.pdf')[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 1 0 1 0 1 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 1 0 0 1 0 0 1 1 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 0 0 0 0 0 0 0 1 0 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
0.9441340782122905
0.8333333333333334
0.9232497634815515
0.8680555555555555
124 FRB20181128C
154 FRB20181218C
203 FRB20181231B
292 FRB20190206A
323 FRB20190218B
454 FRB20190423B
455 FRB20190423B
Name: tns_name, dtype: object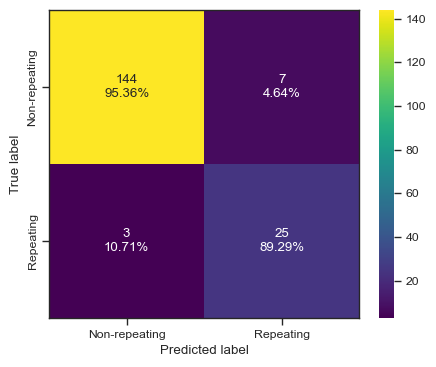
sns.set_theme('paper','ticks')
result = permutation_importance(clf, chime_data, chime_target, n_repeats=100, n_jobs=-1,scoring=f2_score)
forest_importances = pd.Series(result.importances_mean, index=columns_to_use)
fig, ax = plt.subplots()
forest_importances.plot.bar(yerr=result.importances_std, ax=ax,capsize=4)
ax.set_ylabel(r"Mean $F_2$ decrease")
fig.tight_layout()
plt.show()
# fig.savefig('./paper/lgbm_fi.pdf')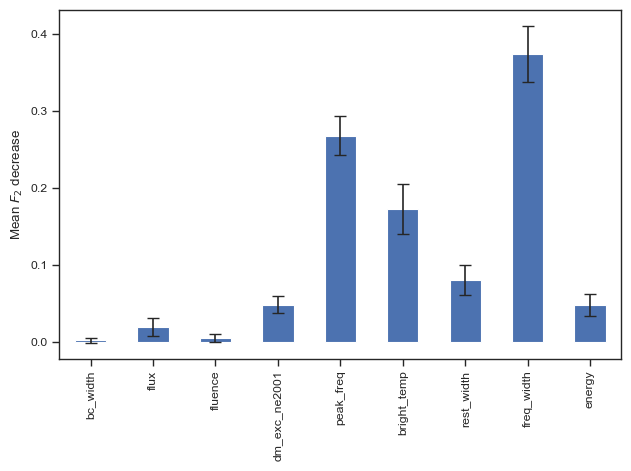
clf = XGBClassifier(use_label_encoder=False)
# clf = CatBoostClassifier()
clf.fit(X, y)
predictions = clf.predict(test_X)
print(predictions)
print(test_y)
print(accuracy_score(test_y,predictions))
print(f1_score(test_y,predictions))
print(roc_auc_score(test_y,predictions))
print(fbeta_score(test_y,predictions,beta=2))
print(CHIME[np.logical_and(clf.predict(chime_data)==1, chime_target==0)]['tns_name'])
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/xgb_cm.pdf')d:\home\lab\sarjana\.venv\lib\site-packages\xgboost\sklearn.py:1421: UserWarning: `use_label_encoder` is deprecated in 1.7.0.
warnings.warn("`use_label_encoder` is deprecated in 1.7.0.")[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 1 0 1 0 1 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 1 0 0 1 0 0 1 1 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0
1 0 0 0 1 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 0 0 0 0 0 0 0 1 0 1 0 1 0 0 0
0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 1 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 1 1
0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0]
0.9385474860335196
0.819672131147541
0.9199385052034058
0.8620689655172414
2 FRB20180729A
124 FRB20181128C
154 FRB20181218C
203 FRB20181231B
232 FRB20190112A
323 FRB20190218B
454 FRB20190423B
455 FRB20190423B
Name: tns_name, dtype: object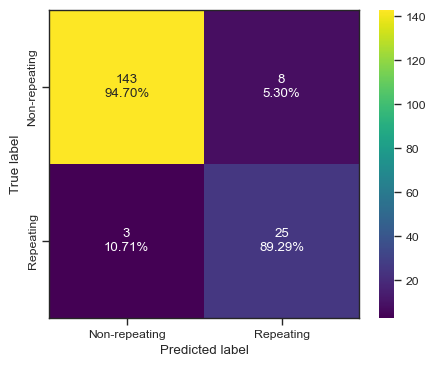
sns.set_theme('paper','ticks')
result = permutation_importance(clf, chime_data, chime_target, n_repeats=100, n_jobs=-1,scoring=f2_score)
forest_importances = pd.Series(result.importances_mean, index=columns_to_use)
fig, ax = plt.subplots()
forest_importances.plot.bar(yerr=result.importances_std, ax=ax,capsize=4)
ax.set_ylabel(r"Mean $F_2$ decrease")
fig.tight_layout()
plt.show()
# fig.savefig('./paper/xgb_fi.pdf')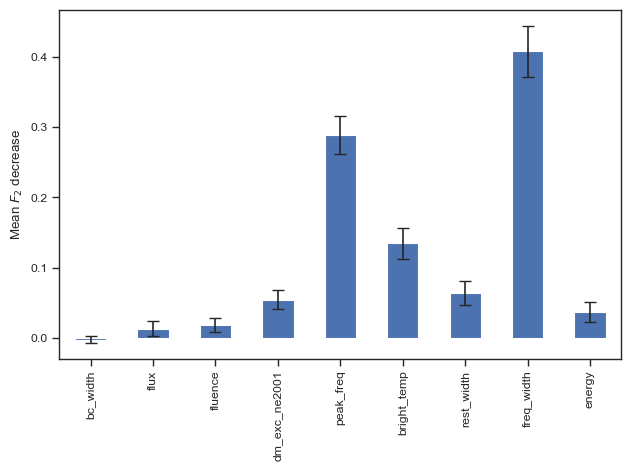
CHIME = load_chime()
columns_to_use = ['bc_width','flux','fluence','dm_exc_ne2001',
'peak_freq',
'bright_temp','rest_width','freq_width','energy']
CHIME['bright_temp'] = np.log10(CHIME['bright_temp'])
CHIME['energy'] = np.log10(CHIME['energy'])
CHIME['rest_width'] = CHIME['rest_width'] * 1000
CHIME['bc_width'] = CHIME['bc_width'] * 1000
CHIME['freq_width'] = np.log10(CHIME['freq_width'])
chime_data = CHIME[columns_to_use]
chime_target = (CHIME['repeater_name'] != '-9999').to_numpy().astype('int')
X,test_X,y,test_y = train_test_split(chime_data,chime_target,test_size=0.3,stratify=chime_target)
X, y = SMOTE().fit_resample(X, y)
clf = DecisionTreeClassifier(max_depth=5)
clf.fit(X, y)
predictions = clf.predict(test_X)
print(predictions)
print(test_y)
print(accuracy_score(test_y,predictions))
print(f1_score(test_y,predictions))
print(roc_auc_score(test_y,predictions))
print(fbeta_score(test_y,predictions,beta=2))
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/tree_cm.pdf')2 78.8 0.00225301 FRB20180729A
12 101.5 0.00225301 FRB20180814A
38 101.0 0.00225301 FRB20180919A
49 94.7 0.00225301 FRB20180928A
75 101.3 0.00225301 FRB20181028A
76 101.3 0.00225301 FRB20181028A
77 101.3 0.00225301 FRB20181028A
78 101.3 0.00225301 FRB20181028A
79 101.3 0.00225301 FRB20181028A
81 62.3 0.00225301 FRB20181030A
82 62.5 0.00225301 FRB20181030B
158 83.6 0.00225301 FRB20181220A
174 92.6 0.00225301 FRB20181223C
221 96.1 0.00225301 FRB20190107B
399 100.8 0.00225301 FRB20190329A
459 79.4 0.00225301 FRB20190425A
571 100.7 0.00225301 FRB20190625E
572 100.7 0.00225301 FRB20190625E
573 100.7 0.00225301 FRB20190625E
576 101.5 0.00225301 FRB20190626A
[1 0 0 0 0 0 1 0 0 0 1 0 0 0 1 0 0 0 0 0 1 1 0 1 1 0 0 0 1 0 1 0 0 1 0 0 1
0 0 0 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 0 1 0 0 0 0
0 0 0 1 1 0 0 0 0 0 1 0 0 0 1 1 0 1 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 1 0 0 0
0 0 1 0 1 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 1 0 0 1 0 1 0 0 0 0 0 0 0 0 0 0 1
0 0 0 0 1 0 0 0 0 1 1 0 1 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 0 1 0]
[0 0 0 0 0 0 0 0 0 0 1 0 0 0 1 1 0 0 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 1 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 1 0 0 0 0
0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 1 0 1 0 0 1 1 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 0 0 0 0 0 1
0 0 0 0 1 0 0 0 0 1 0 0 1 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 0 1 0]
0.8715083798882681
0.676056338028169
0.8656575212866604
0.7741935483870968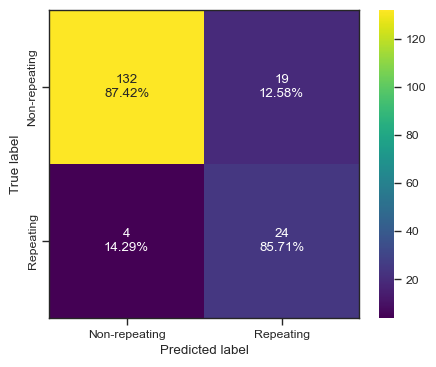
viz = dtreeviz(clf, chime_data, chime_target,feature_names=columns_to_use,class_names=['Non-repeating','Repeating'],label_fontsize=18,ticks_fontsize=10)
viz
# viz.save('./paper/tree_vis_new.svg')d:\home\lab\sarjana\.venv\lib\site-packages\sklearn\base.py:450: UserWarning: X does not have valid feature names, but DecisionTreeClassifier was fitted with feature names
warnings.warn(CHIME = load_chime()
CHIME['bright_temp'] = np.log10(CHIME['bright_temp'])
CHIME['freq_width'] = np.log10(CHIME['freq_width'])
clf = imbpipeline(steps = [['scaler', StandardScaler()],
['smote', SMOTE()],
['classifier', DecisionTreeClassifier(max_depth=3)]])
d2_columns = ['bright_temp','freq_width']
chime_data_2d = CHIME[d2_columns]
X_2d,test_X_2d,y_2d,test_y_2d = train_test_split(chime_data_2d,chime_target,test_size=0.3,stratify=chime_target)
clf.fit(X_2d, y_2d)
predictions = clf.predict(test_X_2d)
print(predictions)
print(test_y_2d)
print(accuracy_score(test_y_2d,predictions))
print(f1_score(test_y_2d,predictions))
print(roc_auc_score(test_y_2d,predictions))
print(fbeta_score(test_y_2d,predictions,beta=2))
categories = ['Non-repeating', 'Repeating']
cf = confusion_matrix(test_y_2d, predictions)
make_confusion_matrix(cf,
categories=categories,
cmap=plt.cm.viridis,
figsize=(5,4),
sum_stats=None)
# plt.savefig('./paper/2d_confusion.pdf')
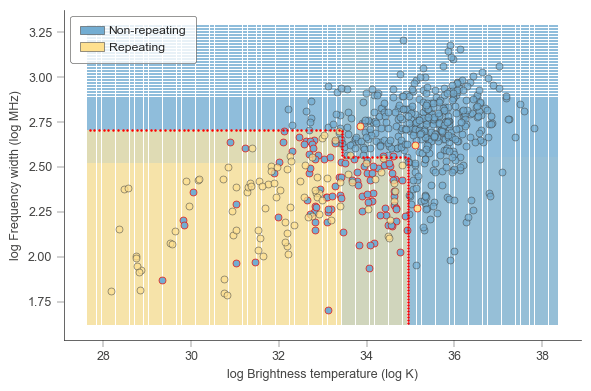
fig,ax = plt.subplots(figsize=(6,4))
clfviz(clf, chime_data_2d, chime_target, ax=ax, ntiles=100,
show=['instances', 'boundaries', 'probabilities', 'misclassified'],
class_names=['non-repeating','repeating'],
feature_names=['log Brightness temperature (log K)','log Frequency width (log MHz)'],
colors={'class_boundary': 'red','classes':[None, None, ["#73ADD2", "#FEE08F"]]})
box1 = patches.Rectangle((0, 0), 20, 10, linewidth=.4, label='Non-repeating',edgecolor='#444443',facecolor='#73ADD2')
box2 = patches.Rectangle((0, 0), 20, 10, linewidth=.4, label='Repeating',edgecolor='#444443',facecolor='#FEE08F')
boxes = [box1,box2]
leg = ax.legend(handles=boxes,frameon=True,shadow=False,fancybox=True,handletextpad=.35,borderpad=.8,edgecolor='#444443')
leg.get_frame().set_linewidth(.5)
plt.tight_layout()
plt.show()
# fig.savefig('./paper/2d_boundary.pdf')2 78.8 0.00225301 FRB20180729A
12 101.5 0.00225301 FRB20180814A
38 101.0 0.00225301 FRB20180919A
49 94.7 0.00225301 FRB20180928A
75 101.3 0.00225301 FRB20181028A
76 101.3 0.00225301 FRB20181028A
77 101.3 0.00225301 FRB20181028A
78 101.3 0.00225301 FRB20181028A
79 101.3 0.00225301 FRB20181028A
81 62.3 0.00225301 FRB20181030A
82 62.5 0.00225301 FRB20181030B
158 83.6 0.00225301 FRB20181220A
174 92.6 0.00225301 FRB20181223C
221 96.1 0.00225301 FRB20190107B
399 100.8 0.00225301 FRB20190329A
459 79.4 0.00225301 FRB20190425A
571 100.7 0.00225301 FRB20190625E
572 100.7 0.00225301 FRB20190625E
573 100.7 0.00225301 FRB20190625E
576 101.5 0.00225301 FRB20190626A
[0 1 1 1 0 0 1 0 0 1 0 0 0 0 1 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0
0 0 0 0 0 1 0 0 1 1 1 0 1 1 1 0 1 0 0 0 1 0 1 1 0 1 1 1 0 0 0 0 0 0 0 1 1
0 0 0 0 0 1 1 1 0 1 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 1 0 0 0 1 0
1 0 1 1 0 0 0 1 1 1 0 1 1 0 0 0 1 0 1 0 0 0 0 0 0 1 0 1 1 0 0 0 0 1 1 0 0
1 0 0 0 1 0 0 1 1 0 0 1 0 0 0 0 1 1 1 1 0 1 0 0 0 1 0 0 0 0 0]
[0 0 0 1 0 0 0 0 0 0 0 0 0 0 1 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 1 0 0 1 0 0 0 1 0 0 1 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 1
0 0 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 1 1 0 0 0 1 1 0 0 1 1 0 0 0 1 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0
1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 1 0 0 0 0 0 0 0 0 0 0 0]
0.8100558659217877
0.6136363636363636
0.8728713339640493
0.7848837209302326d:\home\lab\sarjana\.venv\lib\site-packages\sklearn\base.py:450: UserWarning: X does not have valid feature names, but StandardScaler was fitted with feature names
warnings.warn(
d:\home\lab\sarjana\.venv\lib\site-packages\sklearn\base.py:450: UserWarning: X does not have valid feature names, but StandardScaler was fitted with feature names
warnings.warn(